Coleoptera
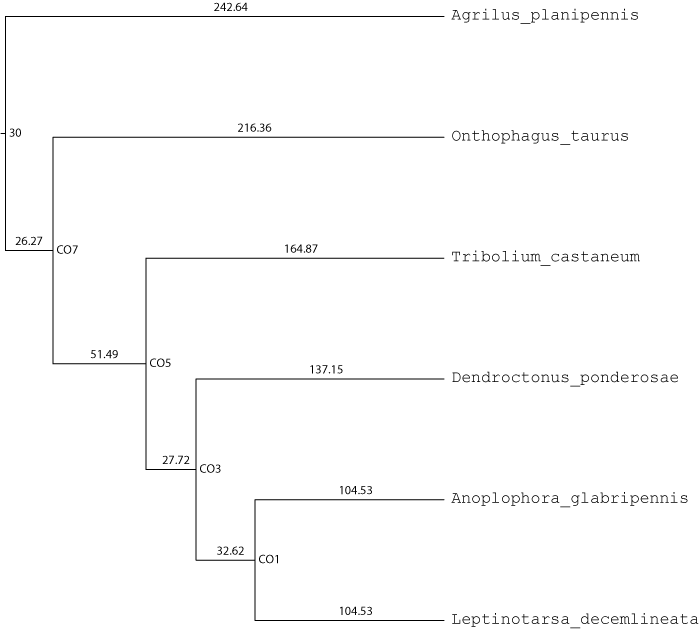
| # species | 6 |
| # single-copy orthologs | 3932 |
| Fossil calibrations | Node 30 at 208.5-411mya |
| # gene families for CAFE | 11598 |
| CAFE estimated lambda | 0.0010 |
Orthogroups were obtained from OrthoDBv8. Single copy sequences were aligned with PASTA and RAxML was used to make gene trees. ASTRAL was used to estimate the species tree topology from gene trees while RAxML on a concatenated alignment was used to estimate branch lengths in terms of relative substitutions. r8s was used with the fossil calibrations to get branch lengths in terms of millions of years. This species tree was then used with CAFE and Dupliphy to reconstruct ancestral gene counts and gains and losses were assessed.
